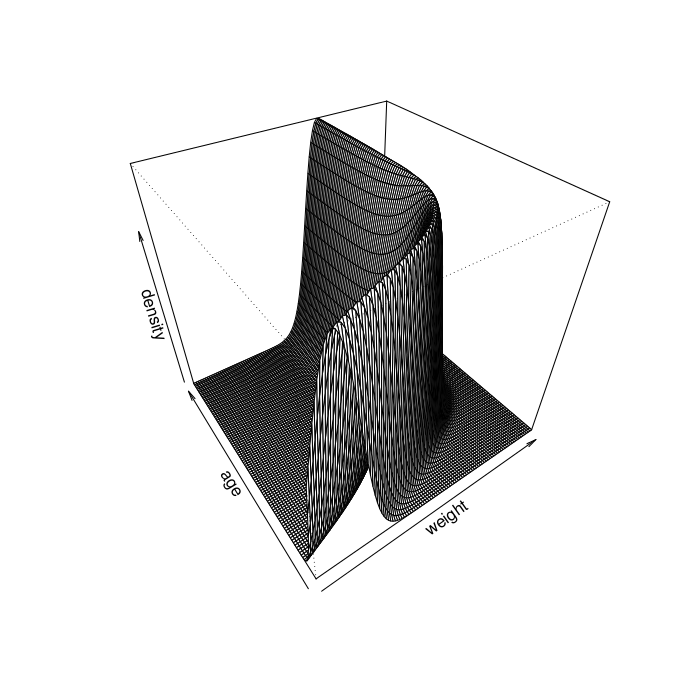
Gompertz Model Visualization
# Gomperz growth function gomp <- function(x, a, b, k) a*exp(-b*exp(-k*x)) # Normal model with Gompertz mean function likelihood <- function(weight, age, sigma, a, b, k) { mu <- gomp(age, a, b, k) dnorm(weight, mu, sigma) } # Visualize the model visualize <- function(phi=40, theta=-35) { weight <- seq(0, 250, length.out=100) age <- seq(0, 50, length.out=100) dens <- outer(weight, age, likelihood, sigma=20, a=170, b=2, k=0.21) persp(weight, age, dens, phi=phi, theta=theta, xlab="weight", ylab="age", zlab="density") }
Web Presentation for Data Frames
I know there is some functionality for this in the Hmisc and R2HTML packages. Can you get alternating row colors with the functions in these packages?
| Murder | Assault | UrbanPop | Rape | |
| Alabama | 13.2 | 236 | 58 | 21.2 |
| Alaska | 10 | 263 | 48 | 44.5 |
| Arizona | 8.1 | 294 | 80 | 31 |
| Arkansas | 8.8 | 190 | 50 | 19.5 |
| California | 9 | 276 | 91 | 40.6 |
| Colorado | 7.9 | 204 | 78 | 38.7 |
# Try: # data(USArrests) # webpage(head(USArrests)) webpage <- function(object, ...) UseMethod("webpage") HEADER <- " <!DOCTYPE html> <html><head> <meta http-equiv=\"Content-Type\" content=\"text/html;charset=utf-8\" /> <style type=\"text/css\"> table { border: 0px; padding: 0px; } tr.even { background: #E2EBF0; text-align: right; } tr.odd { background: #FFFFFF; text-align: right; } tr.name { background: #1F2D49; color: white; text-align: center; } td.name { background: #1F2D49; color: white; text-align: left; } </style></head><body> " FOOTER <- " </body></html> " webpage.data.frame <- function(object, header=HEADER, footer=FOOTER, ...) { esc <- function (text) { text <- gsub("&", "&", text) text <- gsub("\"", """, text) text <- gsub("'", "'", text) text <- gsub(">", ">", text) gsub("<", "<", text) } row_count <- nrow(object) col_count <- ncol(object) row_names <- gsub(" ", " ", row.names(object)) col_names <- gsub(" ", " ", names(object)) cat(header, "<table><tr class=\"name\"><td class=\"name\"></td><td>", paste(esc(col_names), collapse="</td><td>"), "</td></tr>", sep="") evenodd <- "even" for(i in 1:row_count) { cat("<tr class=\"",evenodd,"\"><td class=\"name\">", esc(row_names[i]), "<td>", paste(esc(format(object[i,], ...)), collapse = "</td><td>"), "</td></tr>", sep="") evenodd <- ifelse(evenodd=="even", "odd", "even") } cat("</table>",footer) }
Compress and Upload Files
I was surprised to find little information regarding compressed file uploads in the PHP / JavaScript literature. The function below serves this purpose (but may not be fault tolerant). It would be cool to use this function in conjunction with a local HTTP server (running in R) to provide a web interface to compress and upload files to remote servers. This function assumes that the remote server has a mechanism to receive the data. I've included a server-side CGI shell script below that simply writes the (compressed) data to disk. Alternatively, one could set up a server-side R script, using rApache to simultaneously receive, decompress, and store the data.
# This function compresses a file using 'xz -9' compression # and uploads the file to a server using the HTTP POST method. # 'packpost' is shorthand for 'compress and upload'. The receiving # server should be set up to receive this upload using a server-side # scripting mechanism. packpost <- function(file, host="localhost", port="80", location="/", quiet = FALSE, query = URLencode(file)) { if(!is.character(file) || length(file) != 1) stop("'file' must be a character vector of length 1") if(!is.character(host) || length(host) != 1) stop("'host' must be a character vector of length 1") if(!is.character(port) || length(port) != 1) stop("'port' must be a character vector of length 1") if(!is.character(location) || length(location) != 1) stop("'location' must be a character vector of length 1") if(!is.logical(quiet) || length(quiet) != 1) stop("'quiet' must be a logical vector of length 1") if(!is.character(query) || length(query) != 1) stop("'query' must be a character vector of length 1") # pack cfile <- tempfile() fcon <- file(file, open="rb") ccon <- xzfile(cfile, open="wb", compression=9) if(!quiet) cat("packpost: compressing", file, "->", cfile, "\n") while(length(buff <- readBin(fcon, "raw", 1024)) > 0) writeBin(buff, ccon) close(fcon) close(ccon) if(!quiet) cat("packpost: compression ratio:", file.info(file)$size / file.info(cfile)$size, "\n") # post if(!quiet) cat("packpost: uploading", cfile, "\n") location <- paste(URLencode(location), "?", URLencode(query), sep="") header <- paste("POST ", location, " HTTP/1.1\r\n", "Host: ", paste(host, port, sep=":"), "\r\n", "Content-Length: ", file.info(cfile)$size, "\r\n", "Content-Type: application/x-xz\r\n\r\n", sep="") ccon <- file(cfile, open="rb") scon <- socketConnection(host, port, open="w+b", blocking=TRUE) cat(header, file=scon) while(length(buff <- readBin(ccon, "raw", 1024)) > 0) writeBin(buff, scon) response <- readLines(scon, n=1) close(scon) close(ccon) if(!quiet) cat("packpost: removing", cfile, "\n") unlink(cfile) return(response) }
#!/bin/bash # This script would be located in a CGI directory on a remote host. # Note that this script alone may not be safe. In particular, The web # server should be configured to limit the upload size / prevent malicious # uploads. DATAFILE="upload-`date +%Y-%b-%d-%H%M-%N`" # Append '.xz' when the data are xz compressed if [ "application/x-xz" = "${CONTENT_TYPE}" ]; then DATAFILE="${DATAFILE}.xz" fi # POST data come from STDIN cat > ${DATAFILE} # Return control to CGI handler echo -e "\r\n"